Predict marginal cumulative incidences with confidence intervals for a target trial population
Source:R/generics.R, R/trial_sequence.R, R/predict.R
predict_marginal.Rd
This function predicts the marginal cumulative incidences when a target trial population receives either the
treatment or non-treatment at baseline (for an intention-to-treat analysis) or either sustained treatment or
sustained non-treatment (for a per-protocol analysis). The difference between these cumulative incidences is the
estimated causal effect of treatment. Currently, the
predict function only provides marginal intention-to-treat and
per-protocol effects, therefore it is only valid when estimand_type = "ITT" or estimand_type = "PP".
Usage
predict(object, ...)
# S4 method for class 'trial_sequence_ITT'
predict(
object,
newdata,
predict_times,
conf_int = TRUE,
samples = 100,
type = c("cum_inc", "survival")
)
# S4 method for class 'trial_sequence_PP'
predict(
object,
newdata,
predict_times,
conf_int = TRUE,
samples = 100,
type = c("cum_inc", "survival")
)
# S3 method for class 'TE_msm'
predict(
object,
newdata,
predict_times,
conf_int = TRUE,
samples = 100,
type = c("cum_inc", "survival"),
...
)Arguments
- object
Object from
trial_msm()orinitiators()or trial_sequence.- ...
Further arguments passed to or from other methods.
- newdata
Baseline trial data that characterise the target trial population that marginal cumulative incidences or survival probabilities are predicted for.
newdatamust have the same columns and formats of variables as in the fitted marginal structural model specified intrial_msm()orinitiators(). Ifnewdatacontains rows withfollowup_time > 0these will be removed.- predict_times
Specify the follow-up visits/times where the marginal cumulative incidences or survival probabilities are predicted.
- conf_int
Construct the point-wise 95-percent confidence intervals of cumulative incidences for the target trial population under treatment and non-treatment and their differences by simulating the parameters in the marginal structural model from a multivariate normal distribution with the mean equal to the marginal structural model parameter estimates and the variance equal to the estimated robust covariance matrix.
- samples
Number of samples used to construct the simulation-based confidence intervals.
- type
Specify cumulative incidences or survival probabilities to be predicted. Either cumulative incidence (
"cum_inc") or survival probability ("survival").
Value
A list of three data frames containing the cumulative incidences for each of the assigned treatment options (treatment and non-treatment) and the difference between them.
Examples
# Prediction for initiators() or trial_msm() objects -----
# If necessary set the number of `data.table` threads
data.table::setDTthreads(2)
data("te_model_ex")
predicted_ci <- predict(te_model_ex, predict_times = 0:30, samples = 10)
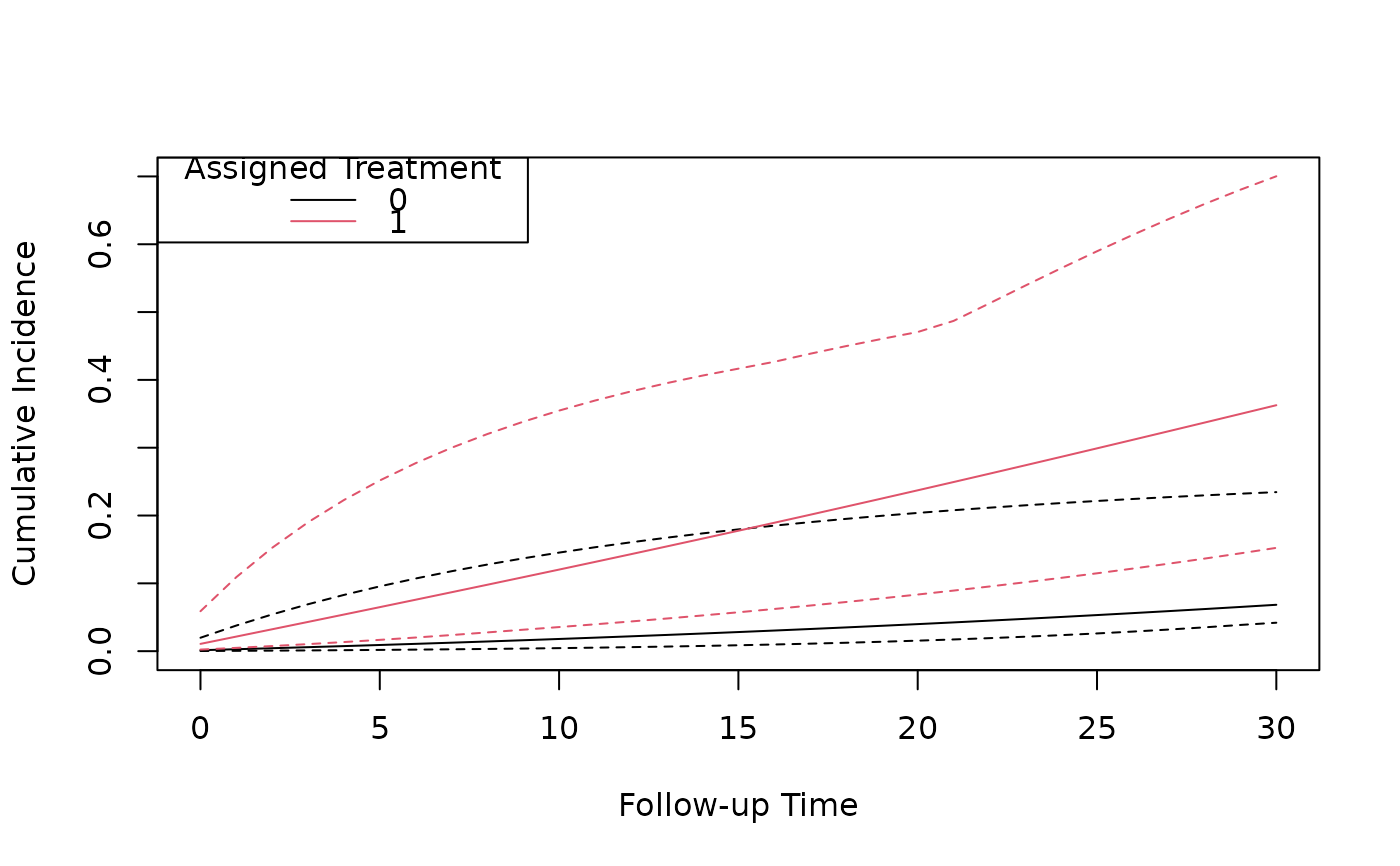
# Plot the cumulative incidence curves under treatment and non-treatment
plot(predicted_ci[[1]]$followup_time, predicted_ci[[1]]$cum_inc,
type = "l",
xlab = "Follow-up Time", ylab = "Cumulative Incidence",
ylim = c(0, 0.7)
)
lines(predicted_ci[[1]]$followup_time, predicted_ci[[1]]$`2.5%`, lty = 2)
lines(predicted_ci[[1]]$followup_time, predicted_ci[[1]]$`97.5%`, lty = 2)
lines(predicted_ci[[2]]$followup_time, predicted_ci[[2]]$cum_inc, type = "l", col = 2)
lines(predicted_ci[[2]]$followup_time, predicted_ci[[2]]$`2.5%`, lty = 2, col = 2)
lines(predicted_ci[[2]]$followup_time, predicted_ci[[2]]$`97.5%`, lty = 2, col = 2)
legend("topleft", title = "Assigned Treatment", legend = c("0", "1"), col = 1:2, lty = 1)
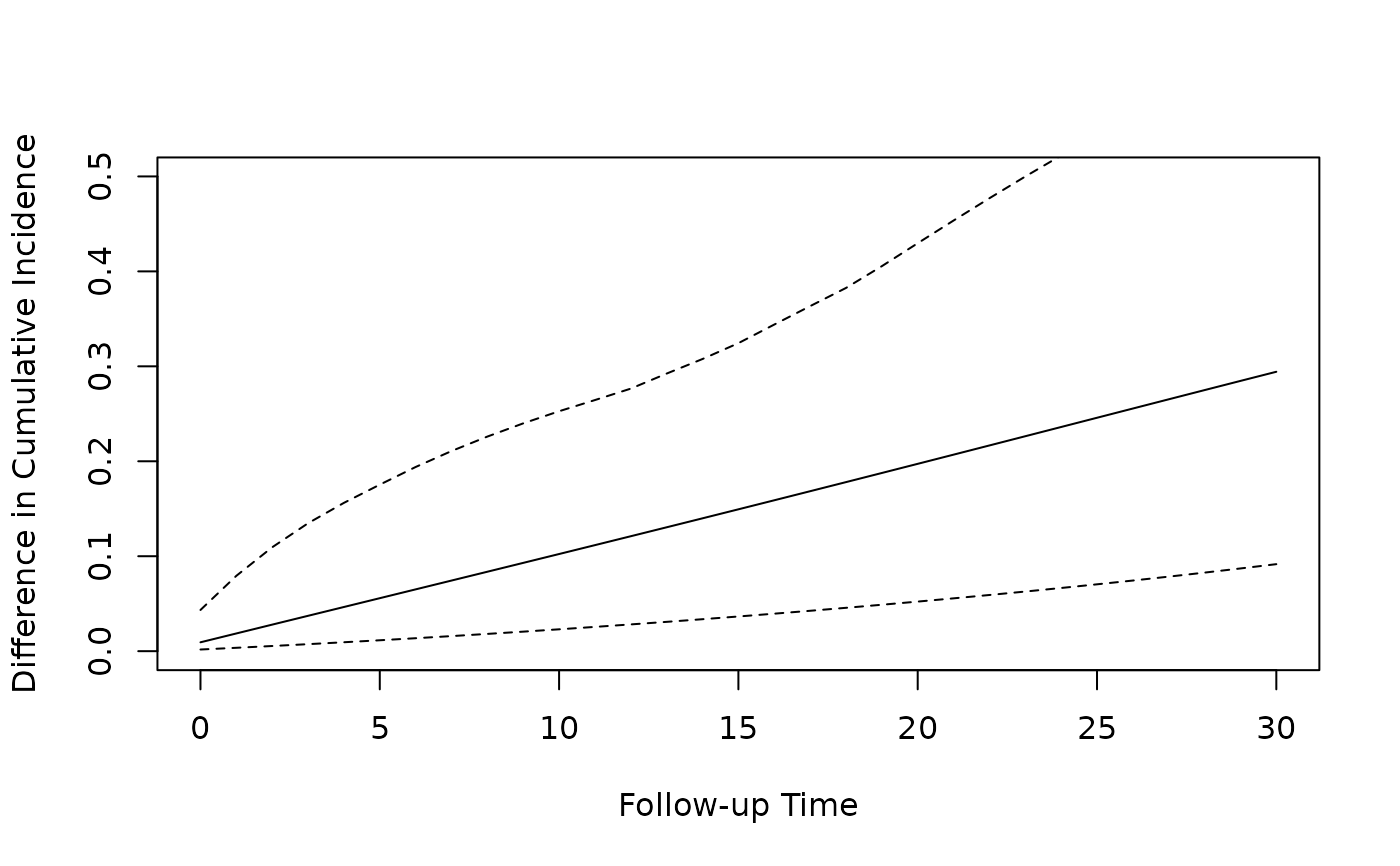
 # Plot the difference in cumulative incidence over follow up
plot(predicted_ci[[3]]$followup_time, predicted_ci[[3]]$cum_inc_diff,
type = "l",
xlab = "Follow-up Time", ylab = "Difference in Cumulative Incidence",
ylim = c(0.0, 0.5)
)
lines(predicted_ci[[3]]$followup_time, predicted_ci[[3]]$`2.5%`, lty = 2)
lines(predicted_ci[[3]]$followup_time, predicted_ci[[3]]$`97.5%`, lty = 2)
# Plot the difference in cumulative incidence over follow up
plot(predicted_ci[[3]]$followup_time, predicted_ci[[3]]$cum_inc_diff,
type = "l",
xlab = "Follow-up Time", ylab = "Difference in Cumulative Incidence",
ylim = c(0.0, 0.5)
)
lines(predicted_ci[[3]]$followup_time, predicted_ci[[3]]$`2.5%`, lty = 2)
lines(predicted_ci[[3]]$followup_time, predicted_ci[[3]]$`97.5%`, lty = 2)